Assistant Professor
Department of Internal Medicine,
Section of Molecular Medicine
Wake Forest School of Medicine
Biswapriya Misra, Ph.D.
Center Affiliations
Address
Email
Center for Biomedical Informatics
Nutrition Research Center (NRC) Building, Rm. G-43
Medical Center Blvd, Winston-Salem, NC 27157
bmisra@wakehealth.edu
Education and Training
B.Sc., Botany (08/2002 )
M.Sc., Botany (07/2004)
Ph.D., Plant Biotechnology (06/2010)
Postdoc, Plant Genomics (2010-2012 )
Postdoc Plant Metabolomics (2013-2016)
Postdoc, Metabolomics (2016-2017)
Sambalpur University, India
Utkal University, India
Indian Institute of Technology Kharagpur, India
Center for Chemical Biology, University of Science Malaysia
University of Florida, Gainesville, USA
Texas Biomedical Research Institute, San Antonio, TX
Prior Positions
Staff Scientist I, Department of Genetics, Texas Biomedical Research Institute, San Antonio, TX, 2017
Research Focus
Metabolomics approaches in diverse biological models and non-model systems toward an integrated analysis of multi-omics datasets
My current work is focused on generating quality metabolomics datasets leveraging the high resolution mass-spectrometry (GC-Orbitrap-MS and LC-Orbitrap-MS) and occasionally NMR datasets to combine with other -omics layers such as genomics (transcriptomics, epigenetics), proteomics, and clinical data sets to provide insights in metabolic changes that are associated with human and non-human primate wellness and disease conditions, specifically metabolic syndromes such as atherosclerosis, diabetes, obesity, and Alzheimer’s disease. In this integrated-/pan-/trans- omics efforts, I intend to weigh in the role of exposures (environmental, dietary, microbial to man-made) as an integral component to these omics efforts for a systems biology understanding of diseases. Using innovative approaches in sample preparation (from challenging cell-/tissue-/ biofluid- types to organs), the power of high resolution tandem mass-spectrometry, ever growing computational and database resources for handling the 'big data', open source resources and reproducible workflows, in conjunction with high dimensional data sets generated at the Center for Precision Medicine (primarily from Professor Olivier and Cox's laboratories) and our collaborators, I intend to identify pathways, networks, and eventually panels of biomarkers that would have relevance to biomedical research and healthcare for identifying targets for therapeutic interventions."
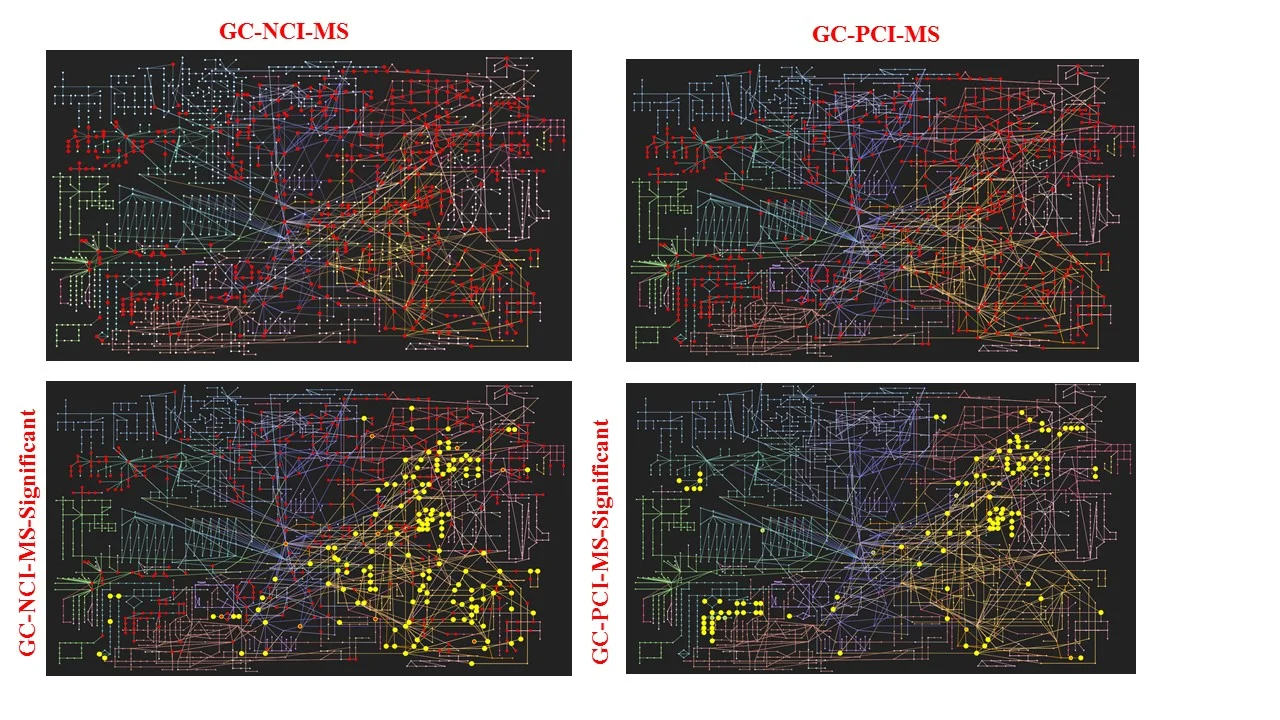
Pathway mapping showing capabilities of a GC-QE-Orbitrap-MS using negative (N) and positive (P) chemical ionization (CI) derivatization strategies from biofluids and tissues for untargeted profiling of metabolites (small organic chemicals).
previous projects
Metabolomics of Single Cell Types: While a Postdoctoral Research Associate at the University of Florida, Gainesville, I worked on an NSF-funded project on Guard Cell Metabolomics to understand leaf cell-type (stomata and mesophyll cells) metabolism in response to biotic and abiotic stress. It involved optimization and building of a targeted HPLC-MRM-MS/MS library of 400+ compounds using a 4000-QTRAP (Sciex) system, and establishment of methods, protocols, and tools for untargeted GC-MS (both Agilent and Thermo Scientific single, tripe-quadrupole and QToF MS instruments), and untargeted profiling using UPLC-ESI-ToF-MS/MS system (Noble Foundation, OK). This work included uni- and multi-variate analyses and visualization, and in house single cell-type technology method development (protoplasting, cell suspensions, sub-cellular sampling), organization and development of workflows based on open source tools, software, databases and computational resources, for metabolomics projects.
Non-Model Organism Genomics:
My postdoctoral work at the Center for Chemical Biology (CCB), University of Science Malaysia USM, Penang, Malaysia, included serving as a Rubber Genome Project Coordinator for the multi-institutional team that reported the first ever rubber tree (Hevea brasiliensis) genome (BMC Genomics). In addition to team-building by coordinating multi-national efforts alongside the PI, I gained experience in genomics (handling of NGS data from multiple platforms), transcriptomics (RNA-seq data, Roche NimbleGen microarrays chip development and validation, WaferGen nano-well qRT-PCR technology), proteomics (label-free MS/MS data analysis), bioinformatics (rubber genome database construction), pathway mapping, and visualization tools. I was involved in large-scale cloning, genome-wide data mining, annotation, curation, wet-lab validation, experimental design, and patent and manuscript drafting associated with the Rubber Genome Project as well as filing of three US patents.